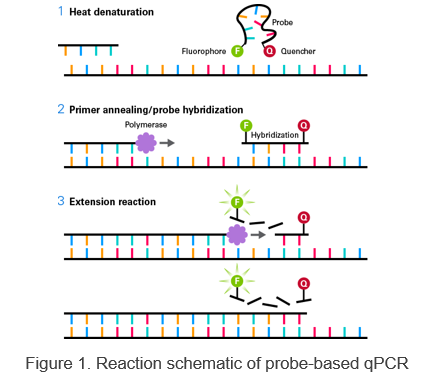
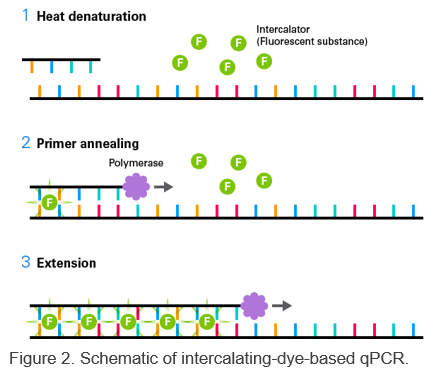
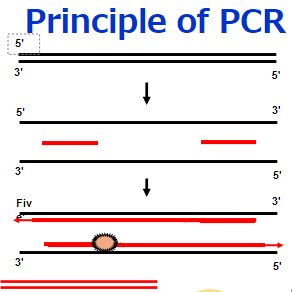
Three-step PCR includes denaturation, annealing, and extension steps. This type of protocol should be used when the Tm of the primers is lower than the extension temperature or is less than 68°C.
If the melting temperature of the primer (Tm) is close to the extension temperature (72°C) or a few degrees lower, consider using a two-step PCR protocol that includes a denaturation step and a combined annealing/extension step. With this protocol, the annealing temperature should not exceed the extension temperature.